What is FISH? FISH is a technique for the detection of specific nucleic acid sequences (DNA or RNA) in the cells or tissues where they reside based on complementary base-pairing of double-stranded nucleic acids. The name of the technique spells out its key utilities:
F: Fluorescence microscopy is used to image target nucleic acid sequence locations. Other variants of this technology use chromogenic labelling (CISH).
IS: Target nucleic acid sequences are detected in situ, in the spatial context of the cells or tissues where they reside. FISH provides information on location, as well as identity and multiplicity of target nucleic acid sequences.
H: The underlying mechanism for recognition of specific nucleic acid sequences is hybridization of single-stranded probe sequences with single-stranded target sequences via complementary base pairing (Figure 1).
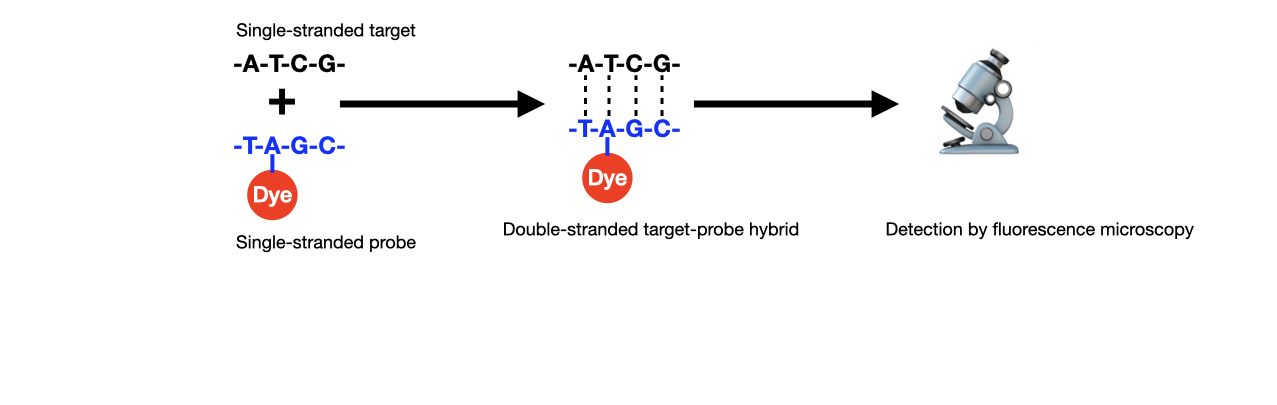
Figure 1. Schematic of fluorescence in situ hybridization (FISH). DNA probes used in clinical cytogenetics are typically around 100,000 bases (100 kb) in length with about 5 dyes per 100 bases (Credit: Lumencor)
FISH in biomedical research
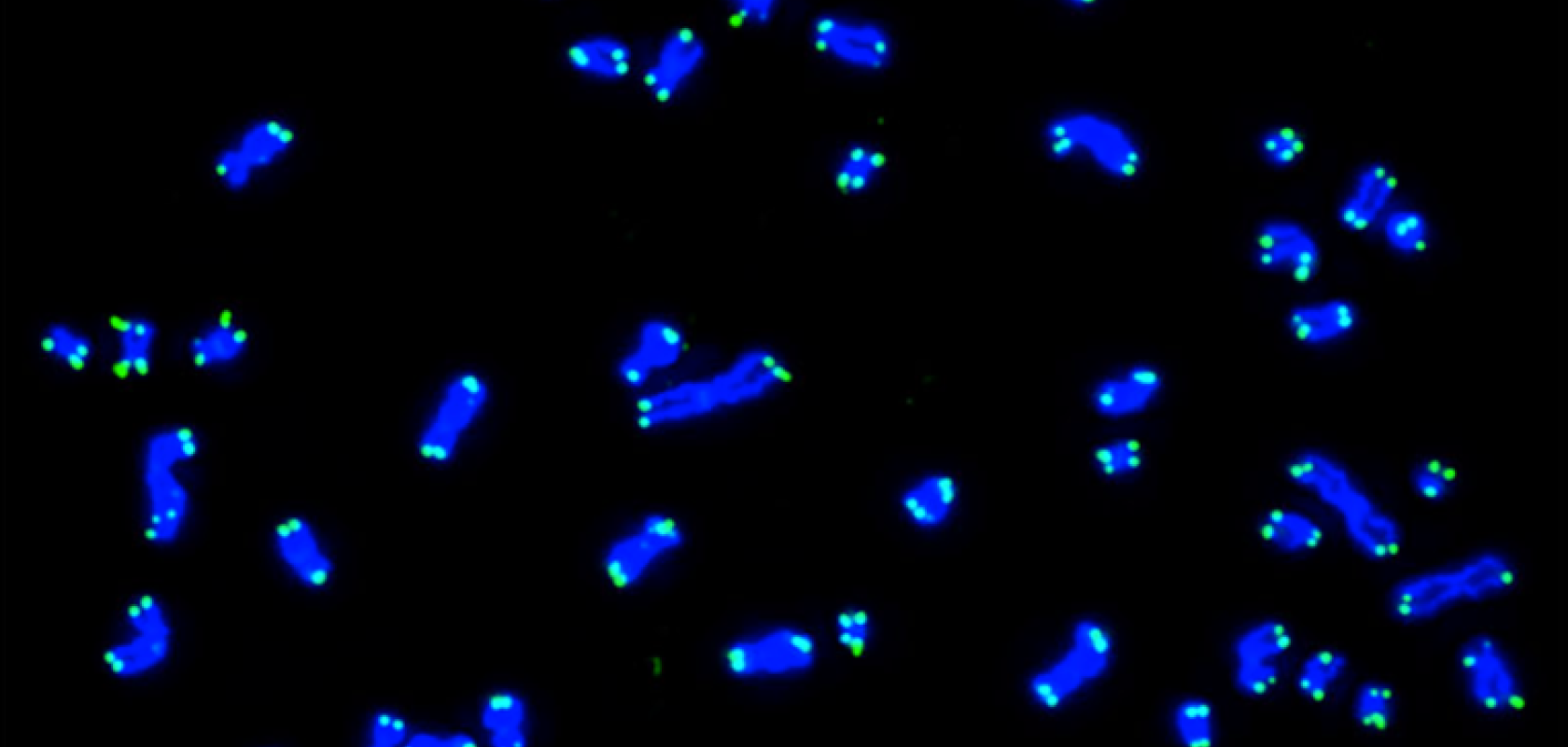
FISH has enabled scientists and clinicians to map genes, study chromosome organisation, diagnose and evaluate genetic diseases, and characterise chromosomal abnormalities. One area of interest has been the analysis of telomeric regions at chromosome ends. Telomeres play a pivotal role in preserving genetic information during cell division by protecting chromosome ends from degradation. Telomere abnormalities such as duplications or deletions can easily be identified via screens that utilise FISH, where telomeres are labelled with fluorescent probe sequences (Figure 2, top image)1. Given that these irregularities can result in various disorders such as age-related syndromes or cancer, the identification of telomere aberrations can provide new insights into disease mechanisms and potentially guide the development of targeted therapies2.
While FISH is a powerful cytogenetic tool used in biomedical research, the visualisation of specific nucleic acid sequences within cells relies heavily on the quality and functionality of the illumination. Through consistent performance in the face of demanding workloads, Lumencor Light Engines enable clinicians, researchers and equipment manufacturers to achieve improved genomic and molecular diagnostics.
Five Illumination requirements for FISH (and how solid-state Light Engines fulfil them)
1. Spectral range to support multiplex detection
The spectral output of Lumencor’s SOLA FISH Light Engine allows fluorescent labels with five or more excitation/emission wavelength ranges to be used to simultaneously detect a corresponding number of different DNA target sequences. Genetic disorders are usually multifactorial making this spectral diversity a ubiquitous requirement. For example, analyte specific reagent kits such as the Vysis MultiVysion PB Multi-color FISH Probe Kit (Abbott Molecular) can be used to simultaneously detect copy numbers of chromosomes 13, 16, 18, 21, and 22, crucial in detecting chromosomal abnormalities significantly impactful on foetal health and viability. Solid-state illuminators such as the SOLA FISH can facilitate engineering of spectral output to optimally align with the requirements of such cytogenetic testing (Figure 3).
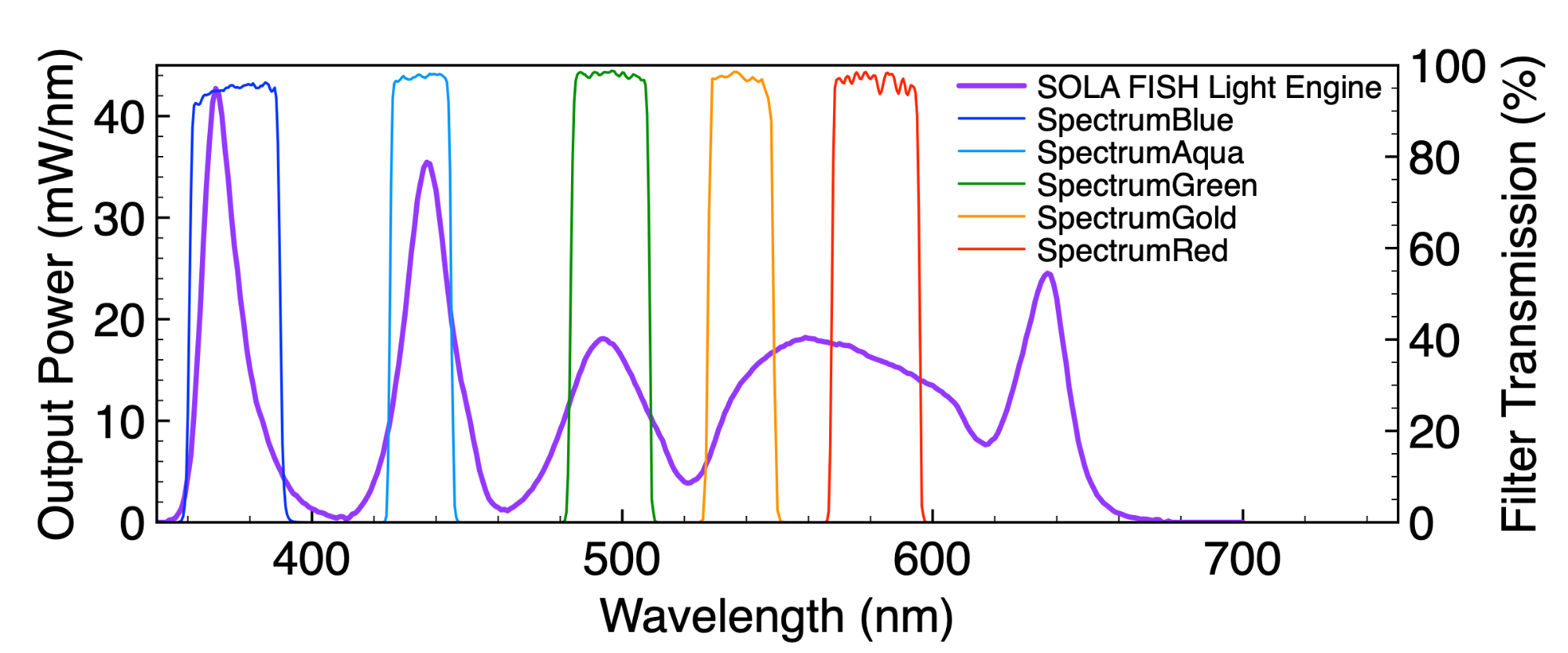
Figure 3. SOLA FISH Light Engine output spectra and Chroma excitation filters for SpectrumBlue, Spectrum Aqua, Spectrum Green, SpectrumGold and SpectrumRed. These fluorophores are incorporated in FISH probes used to determine copy numbers of chromosomes 13, 16, 18, 21, and 22, where multiplexing allows for simultaneous detection. (Credit: Lumencor)
2. Linear intensity adjustment
With solid state illuminators such as the SOLA FISH, light output can be electronically attenuated with a linear control response over a 20-fold dynamic range3. When illumination intensity needs to be adjusted to accommodate variance among specimens, it can be accomplished via electronic software control in a predictable and reproducible manner without the insertion of additional optics such as neutral density filters. Furthermore, the output spectral distribution does not change when electronic attenuation is applied.
3. Unit-to-unit consistency
When purchasing a fleet of illuminators for use in multiple cytogenetic testing facilities, consistency in performance and operational characteristics across all illuminators is crucial. Low variance translates into internally consistent data and operational efficiency. Solid-state Light Engines such as the SOLA FISH demonstrably meet these requirements as illustrated by effectively identical unit-to-unit spectral output (Figure 4)4.
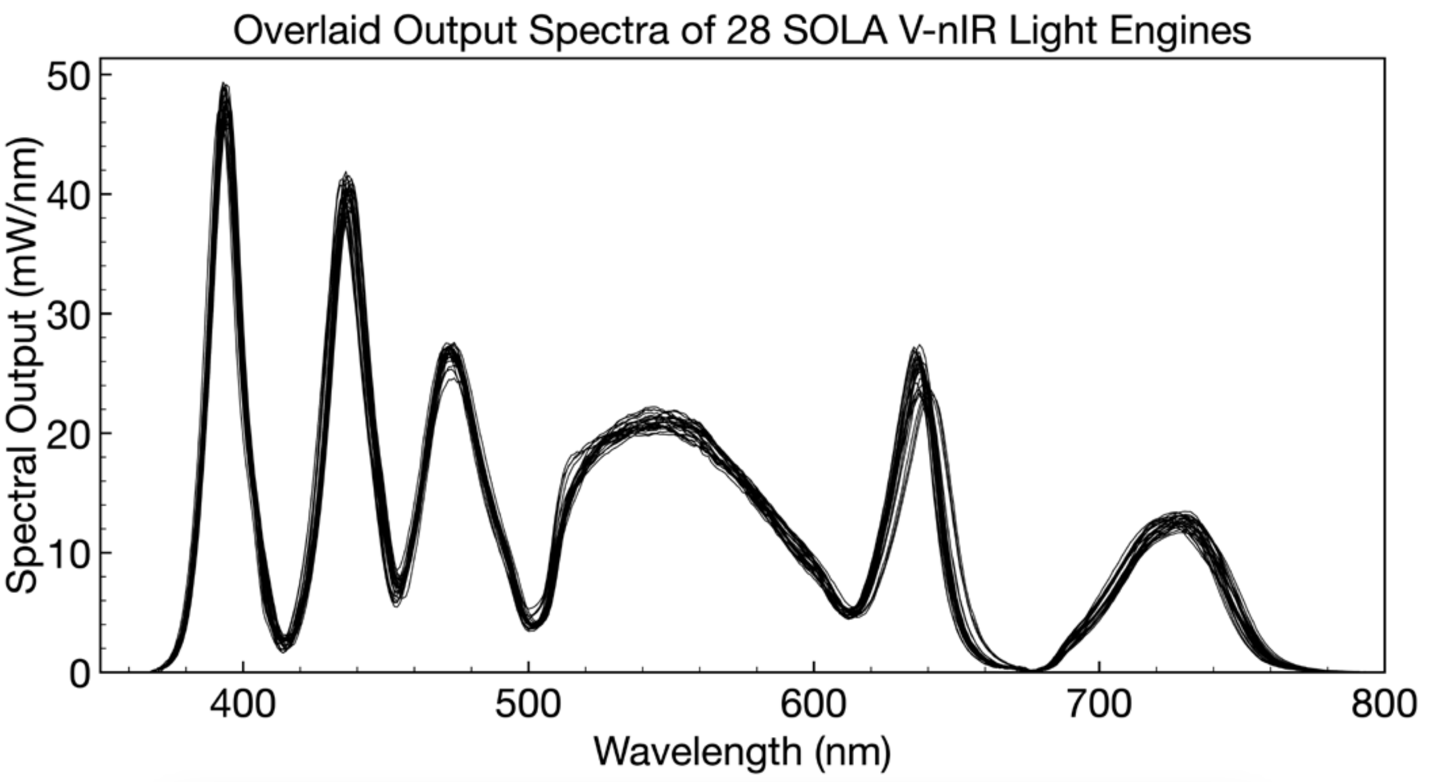
Figure 4. Spectral distribution and optical power of 28 randomly selected Lumencor SOLA Light Engines. A 2% coefficient of variance demonstrates consistency and effectively identical performance between Light Engines. (Credit: Lumencor)
4. Long service lifetime and operational reliability
Solid-state illuminators are maintenance-free, and consumable expenses are minimal compared to mercury and metal halide lamps where expenditure on replacement bulbs can run into thousands of dollars per year. The capital cost of a SOLA FISH can be recouped within two-to-three years of its 10-15 year service lifetime from savings in consumable expenditures5.
5. Built-in metrology and automation compatibility
Modern solid-state illuminators such as the SOLA FISH incorporate on-board microprocessors which monitor system performance in addition to controlling operations. Data such as cumulative light output duration, power consumption and internal operating temperature are available for uploading to laboratory information management systems (LIMS) and supporting regulatory compliance validation. In addition, multiple Light Engines can be linked by ethernet connections, permitting operational control from a single central point.
Overall, Lumencor's SOLA FISH Light Engine combines advanced technology, reliability, and cost-effectiveness, making it the preferred choice for FISH applications in various research and clinical settings.
References
- N Adam, E Degelman, T Beattie et al. (2019) Commun Biol 2:451
- CL Martin, DH Ledbetter (2015) Current Protocols in Human Genetics 84: 8.11.1–8.11.15
- https://lumencor.com/resources/Lumencors-New-Generation-of-Light-Engines
- https://lumencor.com/resources/consistent-light-sources-for-consistent-results
- https://lumencor.com/resources/lumencor-light-engine-stability-and-longevity